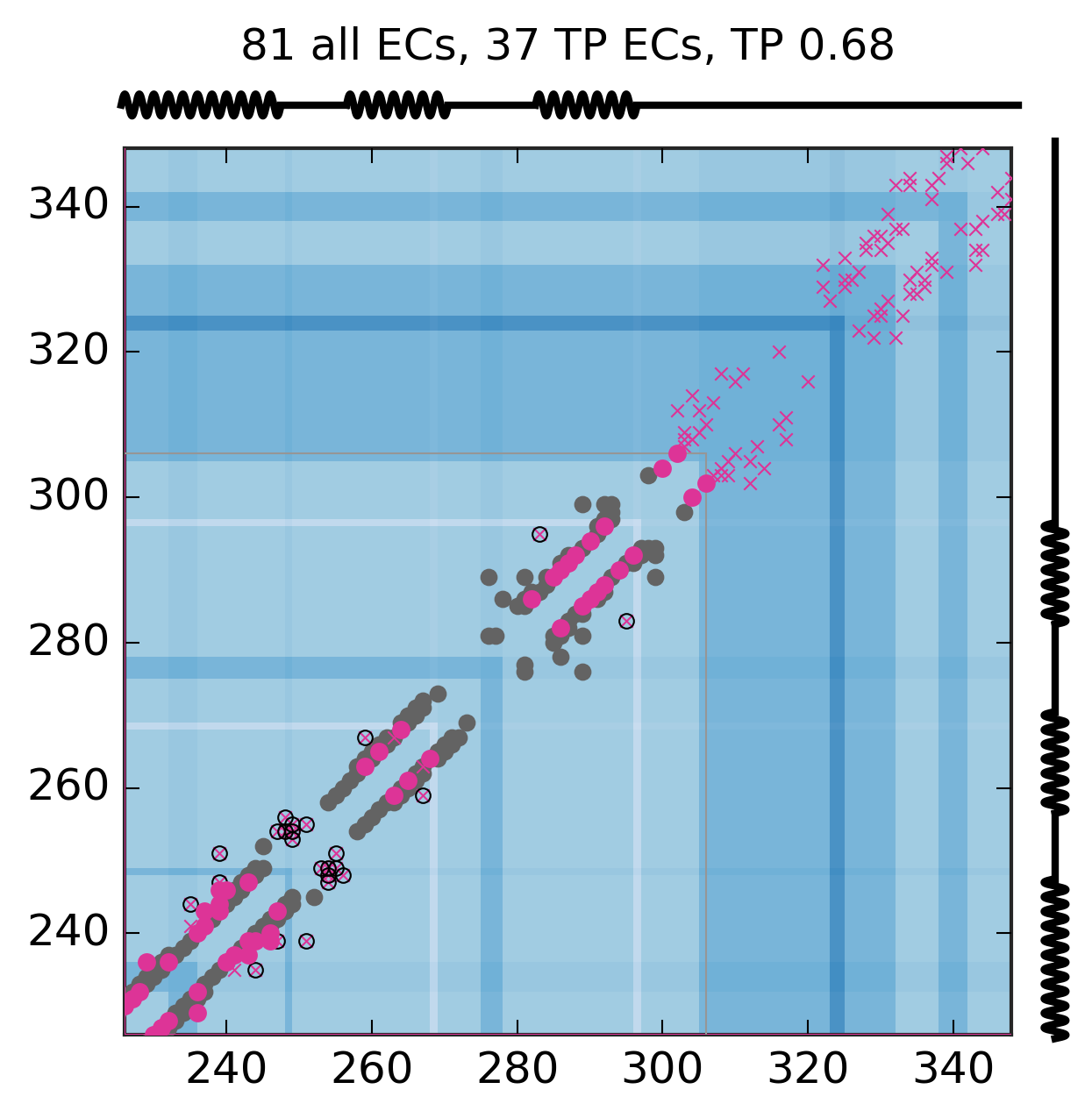
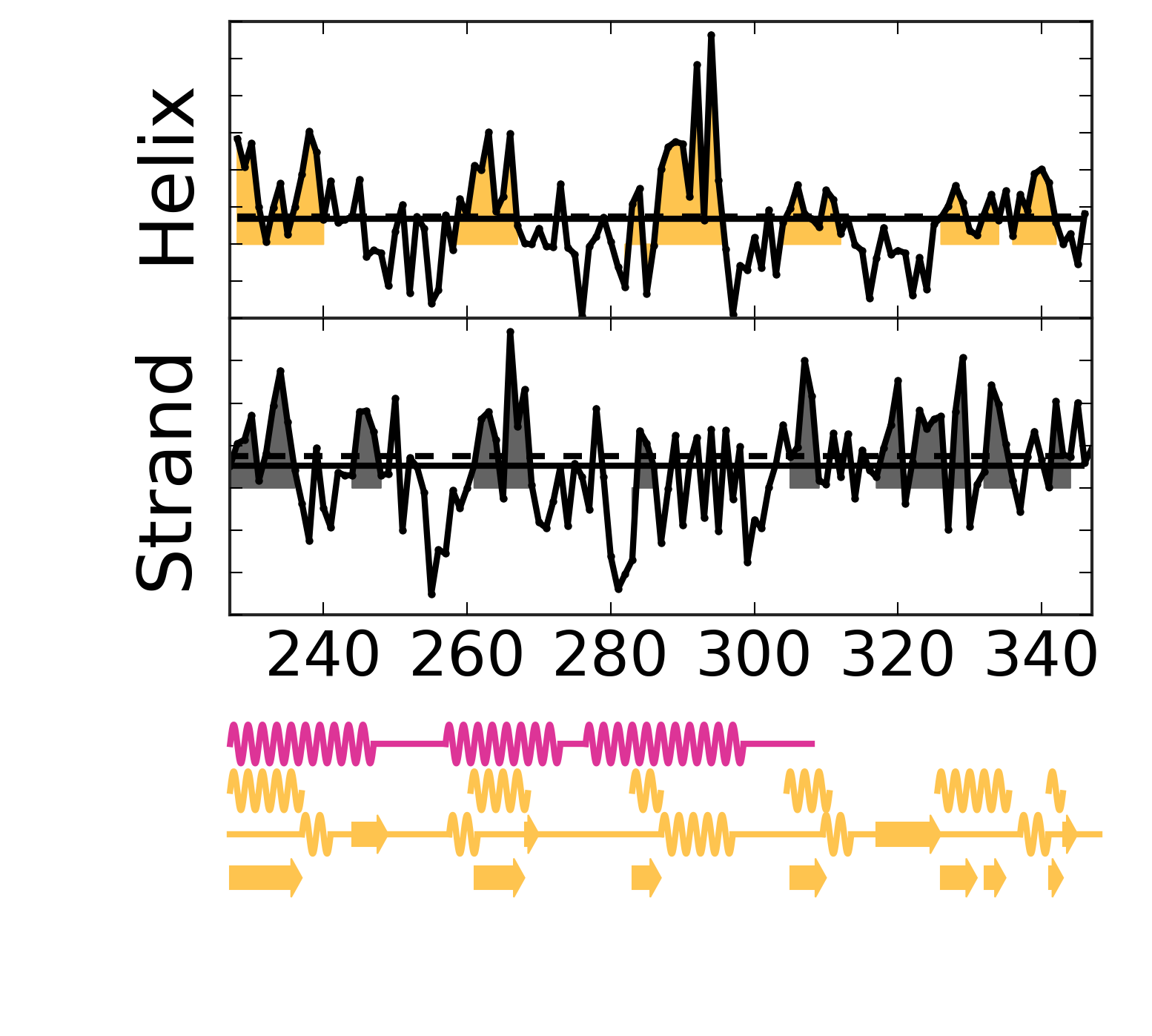
Evolutionary Coupling Analysis
Predicted and experimental contacts
Secondary structure from ECs
Known pdb structures
| pdb |
chain |
| 1P9C |
A |
| 1P9D |
S |
| 1UEL |
B |
| 1YX4 |
A |
| 1YX5 |
A |
| 1YX6 |
A |
| 2KDE |
A |
| 2KDF |
A |
EC score distribution and threshold
Top ECs
| Rank |
Residue 1 |
Amino acid 1 |
Residue 2 |
Amino acid 2 |
EC score |
| 1 |
292 |
Q |
296 |
Q |
0.52 |
| 2 |
290 |
A |
294 |
S |
0.50 |
| 3 |
311 |
S |
317 |
S |
0.39 |
| 4 |
236 |
R |
240 |
A |
0.36 |
| 5 |
264 |
T |
268 |
Q |
0.36 |
| 6 |
326 |
Y |
330 |
Q |
0.36 |
| 7 |
261 |
L |
265 |
I |
0.35 |
| 8 |
325 |
D |
329 |
M |
0.35 |
| 9 |
304 |
E |
308 |
I |
0.33 |
| 10 |
226 |
E |
230 |
R |
0.33 |
| 11 |
300 |
F |
304 |
E |
0.33 |
| 12 |
287 |
I |
291 |
M |
0.31 |
| 13 |
239 |
A |
243 |
A |
0.31 |
| 14 |
239 |
A |
246 |
A |
0.30 |
| 15 |
288 |
A |
292 |
Q |
0.29 |
| 16 |
303 |
A |
307 |
D |
0.29 |
| 17 |
228 |
R |
232 |
E |
0.29 |
| 18 |
323 |
E |
327 |
D |
0.28 |
| 19 |
247 |
G |
254 |
E |
0.28 |
| 20 |
332 |
P |
337 |
S |
0.28 |
| 21 |
304 |
E |
314 |
M |
0.28 |
| 22 |
249 |
A |
255 |
D |
0.28 |
| 23 |
305 |
S |
312 |
S |
0.28 |
| 24 |
325 |
D |
330 |
Q |
0.28 |
| 25 |
333 |
E |
337 |
S |
0.27 |
| 26 |
232 |
E |
236 |
R |
0.27 |
| 27 |
305 |
S |
309 |
D |
0.27 |
| 28 |
325 |
D |
333 |
E |
0.27 |
| 29 |
248 |
I |
256 |
S |
0.27 |
| 30 |
285 |
E |
289 |
Y |
0.27 |
| 31 |
330 |
Q |
336 |
Q |
0.27 |
| 32 |
344 |
G |
348 |
N |
0.27 |
| 33 |
239 |
A |
247 |
G |
0.27 |
| 34 |
339 |
L |
346 |
D |
0.27 |
| 35 |
243 |
A |
247 |
G |
0.27 |
| 36 |
302 |
Q |
306 |
A |
0.26 |
| 37 |
310 |
A |
316 |
T |
0.26 |
| 38 |
303 |
A |
309 |
D |
0.26 |
| 39 |
327 |
D |
331 |
D |
0.26 |
| 40 |
331 |
D |
339 |
L |
0.26 |
| 41 |
239 |
A |
244 |
A |
0.26 |
| 42 |
337 |
S |
341 |
N |
0.26 |
| 43 |
339 |
L |
347 |
P |
0.26 |
| 44 |
249 |
A |
254 |
E |
0.26 |
| 45 |
282 |
T |
286 |
Q |
0.26 |
| 46 |
332 |
P |
343 |
P |
0.26 |
| 47 |
240 |
A |
246 |
A |
0.26 |
| 48 |
331 |
D |
335 |
L |
0.26 |
| 49 |
237 |
R |
241 |
A |
0.25 |
| 50 |
249 |
A |
253 |
T |
0.25 |
| 51 |
251 |
T |
255 |
D |
0.25 |
| 52 |
259 |
A |
267 |
Q |
0.25 |
| 53 |
328 |
V |
334 |
F |
0.25 |
| 54 |
342 |
L |
346 |
D |
0.25 |
| 55 |
286 |
Q |
290 |
A |
0.25 |
| 56 |
227 |
Q |
231 |
Q |
0.25 |
| 57 |
237 |
R |
243 |
A |
0.25 |
| 58 |
329 |
M |
336 |
Q |
0.25 |
| 59 |
330 |
Q |
334 |
F |
0.25 |
| 60 |
229 |
Q |
236 |
R |
0.25 |
| 61 |
308 |
I |
317 |
S |
0.25 |
| 62 |
283 |
E |
295 |
L |
0.24 |
| 63 |
235 |
A |
244 |
A |
0.24 |
| 64 |
316 |
T |
320 |
A |
0.24 |
| 65 |
239 |
A |
251 |
T |
0.24 |
| 66 |
334 |
F |
344 |
G |
0.24 |
| 67 |
259 |
A |
263 |
M |
0.24 |
| 68 |
302 |
Q |
312 |
S |
0.24 |
| 69 |
328 |
V |
335 |
L |
0.24 |
| 70 |
248 |
I |
254 |
E |
0.24 |
| 71 |
337 |
S |
343 |
P |
0.24 |
| 72 |
303 |
A |
308 |
I |
0.24 |
| 73 |
341 |
N |
348 |
N |
0.24 |
| 74 |
235 |
A |
241 |
A |
0.24 |
| 75 |
306 |
A |
310 |
A |
0.23 |
| 76 |
322 |
E |
329 |
M |
0.23 |
| 77 |
307 |
D |
313 |
A |
0.23 |
| 78 |
338 |
V |
344 |
G |
0.23 |
| 79 |
322 |
E |
332 |
P |
0.23 |
| 80 |
263 |
M |
267 |
Q |
0.23 |
| 81 |
334 |
F |
343 |
P |
0.23 |
Alignment robustness analysis
First most common residue correlation
Second most common residue correlation